Artificial cell division a step closer to reality
Researchers at TU Delft have succeeded in replicating a biological mechanism that is essential for cell division in bacteria in the lab. The research is an important step within a larger, ambitious project with the ultimate goal of creating a fully artificial cell that can sustain and divide itself. The researchers have published their findings in Nature Communications.
Even in relatively simple bacterial cells, division is a highly complex process. First, the bacterial DNA has to be copied. Next, proteins, the 'workhorses' of the cell, transport copies of the DNA to the two poles of the cell. Then, in the middle of the cell, a ring is formed that consists of specialised proteins. When the cell is ready to divide, this so-called 'Z-ring' squeezes together, creating two identical daughter cells.
Tag
The proteins that form the Z-ring in most bacteria always do so neatly in the middle of the cell. But how do they know where the middle is? For this, the cell uses other proteins, three in total: MinC, MinD and MinE. Together, they form the 'Min system'. The MinD protein likes to attach itself to the inner cell membrane. The function of the MinE protein is to drive MinD away. And the MinC protein? It is a passenger that hitches a ride with the MinD and MinE proteins.
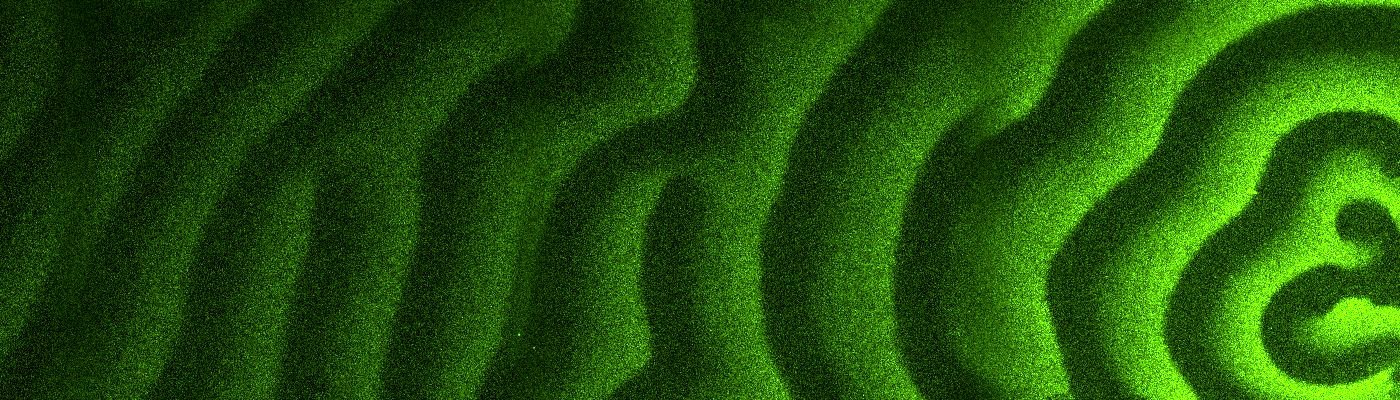
The interplay between these three proteins can be described as a game of tag, in which MinE proteins dislodge MinD from the membrane. Chased away, the MinD proteins seek out a new spot where they can attach themselves to the membrane, only to be chased away once again. This collective behavior creates waves of Min proteins (see picture) that move from one pole of the cell to the other.
These ‘protein waves’ (oscillations) form a gradient in the cell. The highest concentration of Min proteins can be found at the poles, while the protein concentration is the lowest in the centre. This is where the third Min protein, the hitchhiking MinC, comes in. Its function is to suppress the formation of the Z-ring. Since there is only little of it in the middle of the cell, the Z-ring can freely form there.
Synthetic cell
Together with colleagues in The Netherlands and abroad, researchers at TU Delft are trying to build a 'synthetic cell'. The idea is to first reconstruct all the necessary modules separately in the lab, and then to bring them together to form a functional, man-made cell. Division is such a module, and it in turn consists of various mechanisms that work together, one of those being the Min system.
The group of Christophe Danelon at TU Delft has now succeeded in reproducing the Min-system in artificially created vesicles, so-called 'liposomes'. All three proteins were directly expressed from their genes within such liposomes, a process known as ‘cell-free protein synthesis’. "But it is not enough to just insert the DNA of these three genes into a liposome," lead researcher Elisa Godino explains. She compares creating the right environment for the genes to cooking. "You need to have a certain amount of each protein, and the proteins only do their job under the right conditions. A lot of time and research has gone into fine-tuning our recipe."
Ultimately, the researchers managed to reconstruct the Min system in a liposome. Through her microscope, Elisa Godino saw the characteristic 'protein waves' that indicated the system worked. The next step is to reconstruct the mechanism that is responsible for building the aforementioned Z-ring. “We have already confirmed that the Min system we built properly interacts with the core components of the Z-ring”, says Godino.
In the near future, the researchers hope to combine the two cell division mechanisms, which will undoubtedly pose a number of new challenges. But still: step by step, the synthetic cell is becoming a reality.
****
Elisa Godino, Jonás Noguera López, David Foschepoth, Céline Cleij, Anne Doerr, Clara Ferrer Castellà, Christophe Danelon, “De novo synthesized Min proteins drive oscillatory liposome deformation and regulate FtsA-FtsZ cytoskeletal patterns”, Nature Communications (2019)
DOI: 10.1038/s41467-019-12932-w
Elisa Godino
E.Godino@tudelft.nl
015-2782984