Have neural networks gone beyond our understanding?
Artificial intelligence has seen an enormous growth spurt with the application of ‘deep neural networks’. Such networks are taking over more and more tasks from humans in a nearly unparalleled manner. But who still understands how they work?
Neural networks are being used increasingly often and with increasing success. For example in driverless cars, medical diagnosis, the board game ‘GO’ or recognising animals in pictures. Rather than programming a computer explicitly for a certain task, researchers let deep neutral networks (DNN) figure out for themselves what the typical characteristics are of the input and tell us what this input means.
This usually concerns big data, and is always about high-dimensional datasets. Because each pixel is a variable, photographs, for instance, easily have tens of thousands of dimensions. Medical data, such as the results of a gene expression test, are high-
dimensional as well.
People understand the input and output of a neural network. But its inner workings are a black box. It feels unsettling to have computers make judgements in a way that we ourselves do not understand.
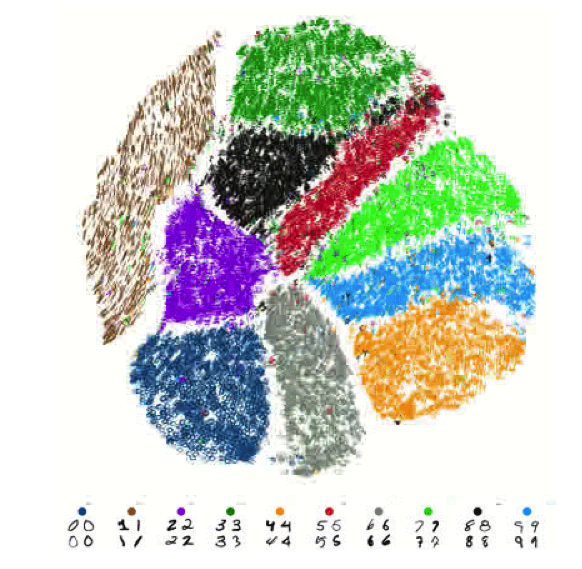
A test dataset of 60,000 hand-written numbers (28x28 pixels), displayed by the tSNE algorithm as ten almost completely unoverlapping clusters.
Improving the algorithm
Nicola Pezzotti, TU Delft doctoral candidate in the Computer Graphics and Visualisation group (Faculty of EEMCS), did a work placement at Google. There, he improved the
algorithm (t-distributed Stochastic Neighbour Embedding, or t-SNE) used to visualise the data in a high- dimensional dataset as a number of low dimensions, suitable for human beings. The algorithm can be used, for instance, to display a large dataset of 60,000 digitalised hand-written numbers (28x28 pixels) as clusters in a two-dimensional figure. In addition, Pezzotti's improved algorithm is a powerful tool for understanding the workings and quality of neural networks.
Visual representation
If a neural network is poorly designed, or poorly trained, it will make mistakes when interpreting new data input. But going through a large validation dataset by hand to separate out unexpected and undesired results would be a hopeless task. After all, the output from a neural network is also extremely high-dimensional as well and hard for people to visualise. In recognising animals, the output for each new figure may comprise, for instance, a long series of numbers, with each number representing the probability of the figure being a dog, pigeon, elephant, etc.
A visual representation of this data, using the tSNE algorithm, gives very quick clarity. Is there too much overlap between clusters, or have some small clusters unexpectedly appeared, perhaps? For computer scientists, these are signs that they need to redesign or retrain the DNN.
Nicola Pezzotti made his implementation of the algorithm so fast that anyone can use it, at home on a simple desktop computer or even via a web browser. In the meantime, Google has added it to its open source software platform for the design and validation of neural networks (‘Tensorflow.js’). The basis for the greatly improved algorithm was to use the physics concept of a ‘field’.
Off-label use
Biomedical researchers at Leiden University Medical Center (LUMC) have applied the tSNE algorithm, but without using a neural network. The researchers measured from large batches of individual cells the extent to which certain proteins appeared on the cell surface.
Visualisation with the tSNE algorithm showed them that undiscovered human immune system cell types and new intermediary stages of stem cells could still be present, and how these could be identified. The researchers were subsequently able to demonstrate the presence of these types of cells by searching for them specifically.