Metabolic engineering of added-value compounds
Metabolic engineering, i.e., the intentional redirection of metabolic fluxes plays an important role in improving yeast strains for a wide range of industrial applications. In contrast to classical methods of strain improvement metabolic engineering has conferred two major advantages: (i) targeted modification of strains without accumulation of unfavorable mutations and (ii) introduction of genes from foreign organisms to equip S. cerevisiae with novel traits. The latter is particularly relevant for industrial biotechnology to provide pathways that extend the spectrum of usable industrial media and/or to produce compounds not naturally formed by S. cerevisiae.
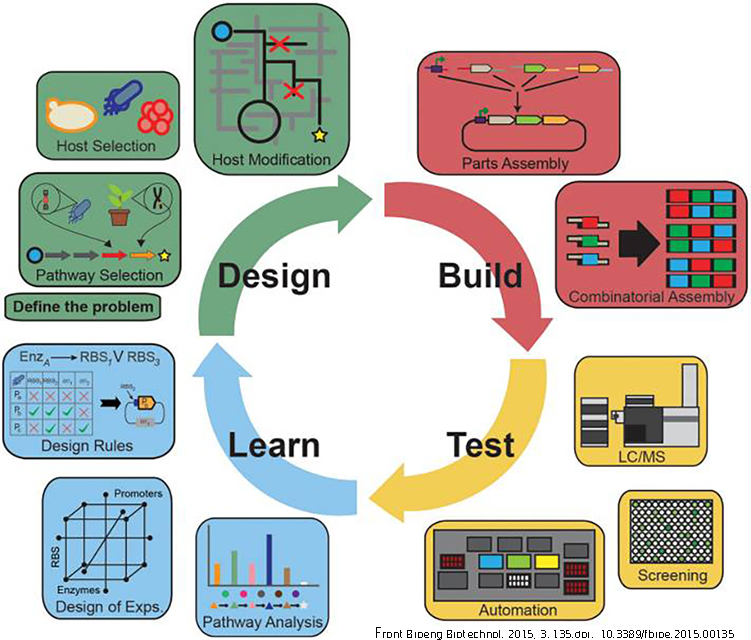
The plant kingdom is enormously rich in bioactive molecules from a wide variety of compound classes (e.g. flavonoids, terpenoids, alkaloids, etc). Only a minute fraction of these molecules are currently applied for manufacturing food ingredients, pharmaceuticals, cosmetics, etc. Systematic exploration of this chemical diversity is complicated by the complexity and costs of both purification of relevant compounds from plant biomass and their de novo chemical synthesis. Industrial microorganisms can be grown at scales ranging from < 0.1 ml (high throughput screening) to > 100 m3 (industrial production). Moreover, industrial microorganisms are unsurpassed in terms of genetic accessibility and detailed knowledge on regulation of their central metabolism, which provides the precursors for product formation.
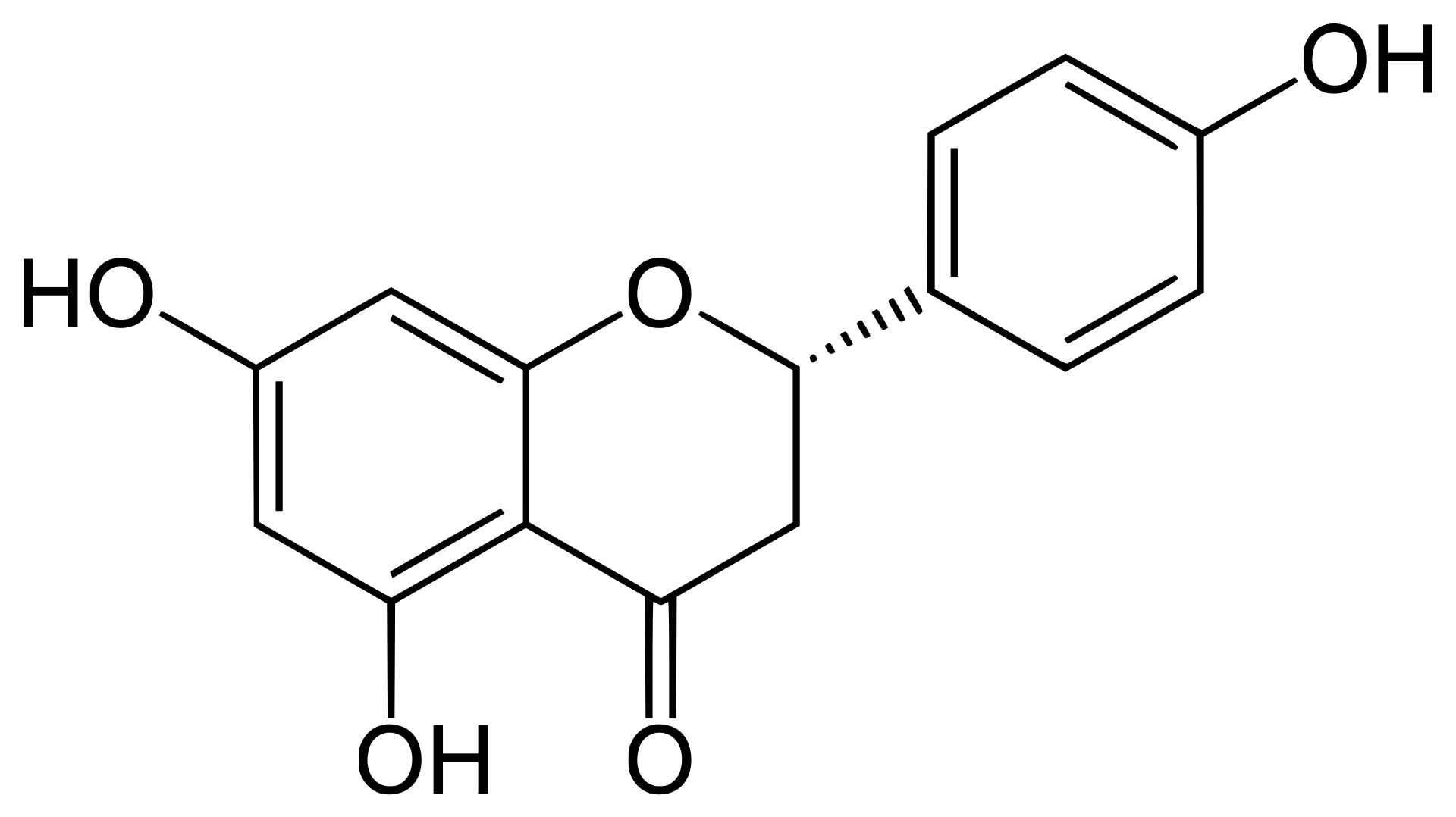
2 Flavonoids comprise a large family of secondary plant metabolic intermediates that exhibit a wide variety of antioxidant and human health-related properties. In an early work, Saccharomyces cerevisiae was engineered to produce the key intermediate flavonoid, naringenin, solely from glucose. For this, specific naringenin biosynthesis genes from Arabidopsis thaliana were selected and introduced in S. cerevisiae. The sole expression of these A. thaliana genes yielded low extracellular naringenin concentrations. To optimize naringenin titers, a yeast chassis strain was developed. Synthesis of aromatic amino acids was deregulated by alleviating feedback inhibition of 3-deoxy-d-arabinose-heptulosonate-7-phosphate synthase (Aro3, Aro4) and byproduct formation was reduced by eliminating phenylpyruvate decarboxylase (Aro10, Pdc5, Pdc6). Together with an increased copy number of the chalcone synthase gene and expression of a heterologous tyrosine ammonia lyase, these modifications resulted in a 80-fold increase of extracellular naringenin titers (to approximately 400 μM) in aerated, pH controlled batch reactors. We demonstrated that S. cerevisiae is capable of de novo production of naringenin by coexpressing the naringenin production genes from A. thaliana and optimization of the flux towards the naringenin pathway. This engineered yeast naringenin production host provides a metabolic chassis for production of a wide range of flavonoids and exploration of their biological functions.
People: Jasmijn Hassing, Mario Beck (start October 2017), research associate position.
2 Schematic representation of the engineered naringenin production pathway in S. cerevisiae. Six A. thaliana genes were overexpressed: PAL1 (phenylalanine ammonia lyase), C4H (Cinnamate 4-hydroxylase), CPR1 (cytochrome P450 reductase), 4CL3 (4-coumaric acid-CoA ligase), CHS3 (chalcone synthase) and CHI1 (chalcone isomerase), and one gene from R. capsulatus; Tal1 (tyrosine ammonia lyase). Dashed lines indicate feedback inhibition. Grey arrows indicate the S. cerevisiae pathway for phenylethanol production. Bold dark grey arrows indicate the naringenin production pathway as described for A. thaliana. Aro3/Aro4: 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase, Pdc1, 5, 6; pyruvate decarboxylases, Aro10; phenylpyruvate decarboxylase.
Funding:


The CHASSY project has received funding from the European Union’s Horizon 2020 Framework Programme for Research and Innovation - Grant Agreement No. 720824. [http://chassy.eu/]


NGI Booster Project Plant Green Synthetic Biology (050-040-211) and Horizon Breakthrough Projects Yeast based valorization of plant metabolic diversity: Engineering Saccharomyces cerevisiae for anthocyanin production (93519011).
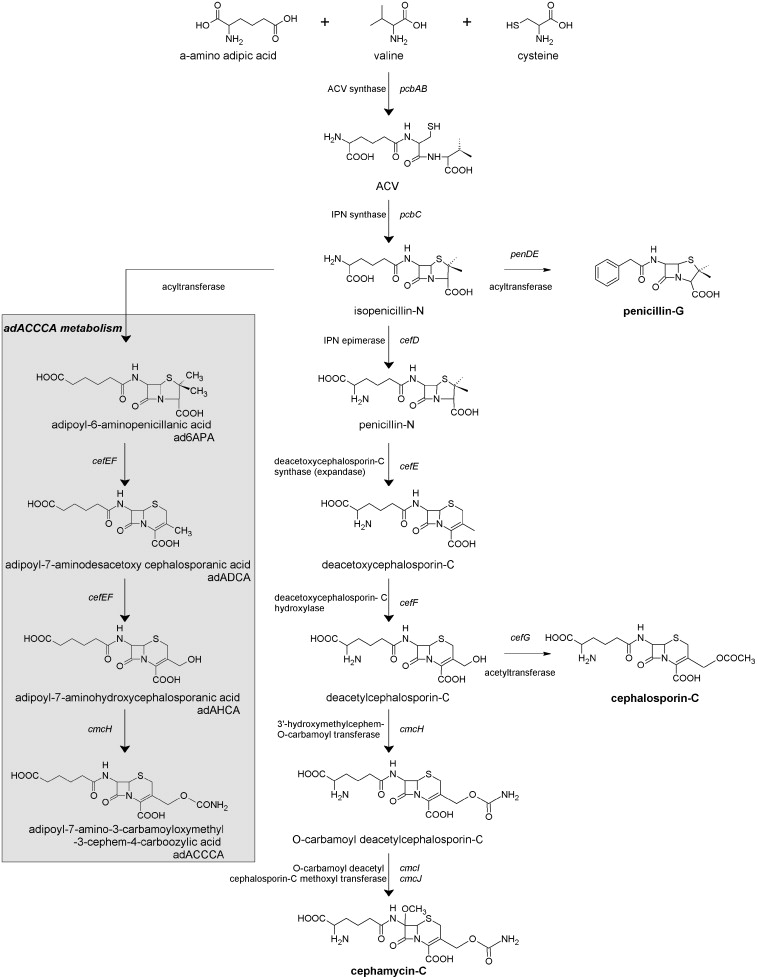
During the past four to five decades, P. chrysogenum strain improvement has been achieved exclusively by classical strain improvement. However robust, this method is time and labor intensive and can be an economic hurdle that limits commercial realization of a promising product or biological process. The availability of the complete genome sequence of P. chrysogenum Wisconsin54-1255 strain, opened new possibilities for research. Molecular basis studies with the implementation of recently developed genetic tools allowed a thorough functional characterization of this important industrial microorganism and to systematically test the involvement of specific genes and pathways on industrial performance. To exemplify this, P. chrysogenum was successfully engineered to produce a novel carbamoylated cephalosporin that can be used as a synthon for semi-synthetic cephalosporin antibiotics. Growth of the engineered strain in the presence of adipic acid resulted in production of adipoyl-7-amino-3-carbamoyloxymethyl-3-cephem-4-carboxylic acid (ad7-ACCCA) and of several adipoylated pathway intermediates. However, only a small fraction of the consumed adipic acid was recovered as ad7-ACCCA. A combinatorial chemostat-based transcriptome study, in which the ad7-ACCCA-producing strain and a strain lacking key genes in beta-lactam synthesis were grown in the presence and absence of adipic acid, enabled the dissection of transcriptional responses to adipic acid per se and to ad7-ACCCA production. Transcriptome analysis revealed that adipate catabolism in P. chrysogenum occurs via beta-oxidation and enabled the identification of genes comprising this pathway. Their functional analyses led to targeted engineering strategies to improve cephalosporin intermediate biosynthesis.
Funding:
IBOS (Integration of Biosynthesis and Organic Synthesis) programme of the Netherlands Organisation for Scientific research (NWO). The project “Transcriptomics and quantitative physiology of β-lactam-producing Penicillium chrysogenum”.

Senternovem project (IS 043055) on the improvement of homologous recombination in Penicillium chrysogenum.
IBOS II (Integration of Biosynthesis and Organic Synthesis II) programme of the Netherlands Organisation for Scientific research (NWO) in collaboration with DSM entitled CellPep (IBOS 053.63.011). “Improving production of -lactams antibiotics by Penicillium chrysogenum – Metabolic engineering based on transcriptome analysis“.

B-Basic (financially supported by the Netherlands Ministry of Economic Affairs and the B-Basic partner organizations (www.b-basic.nl) through, a public-private NWO-ACTS program (ACTS: Advanced Chemical Technologies for Sustainability) “Improving production of semi synthetic cephalosporin antibiotics by Penicillium chrysogenum”.