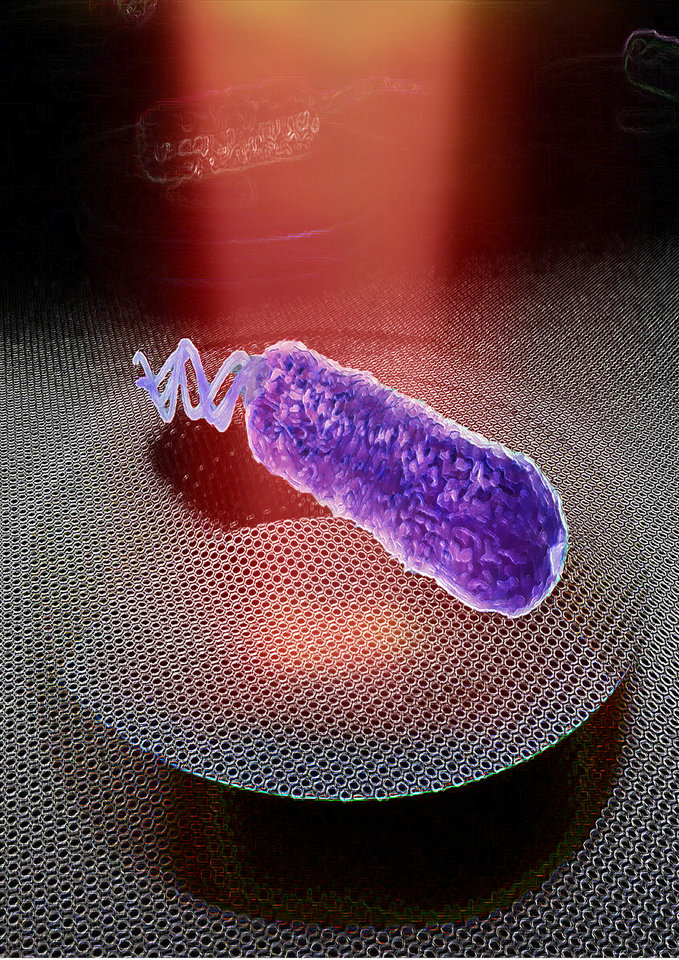
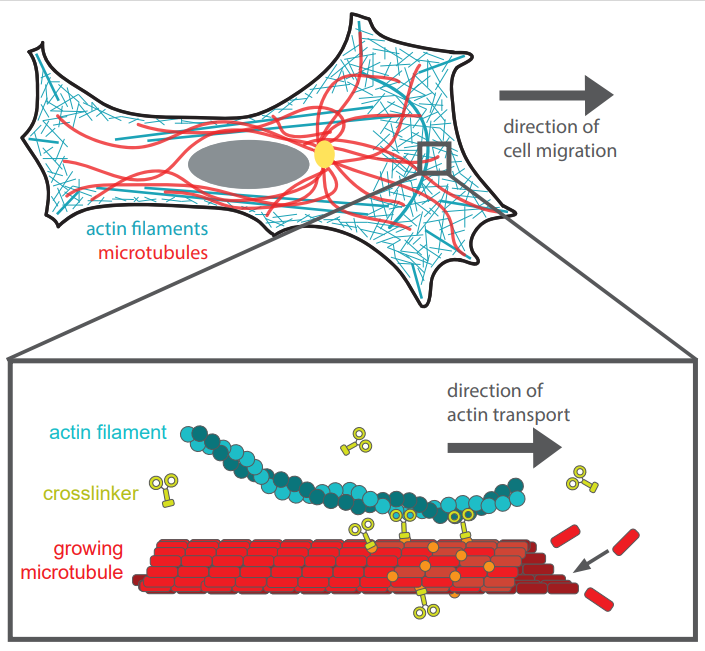
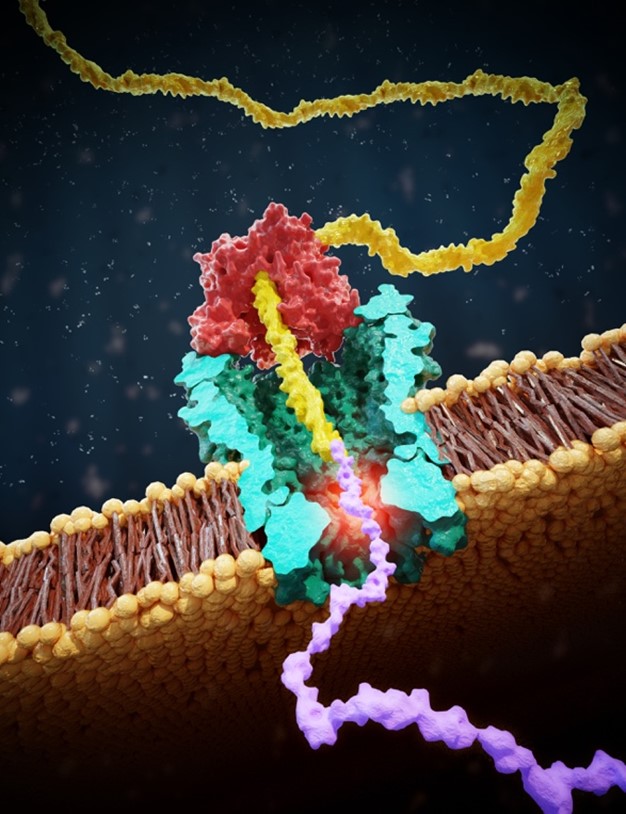
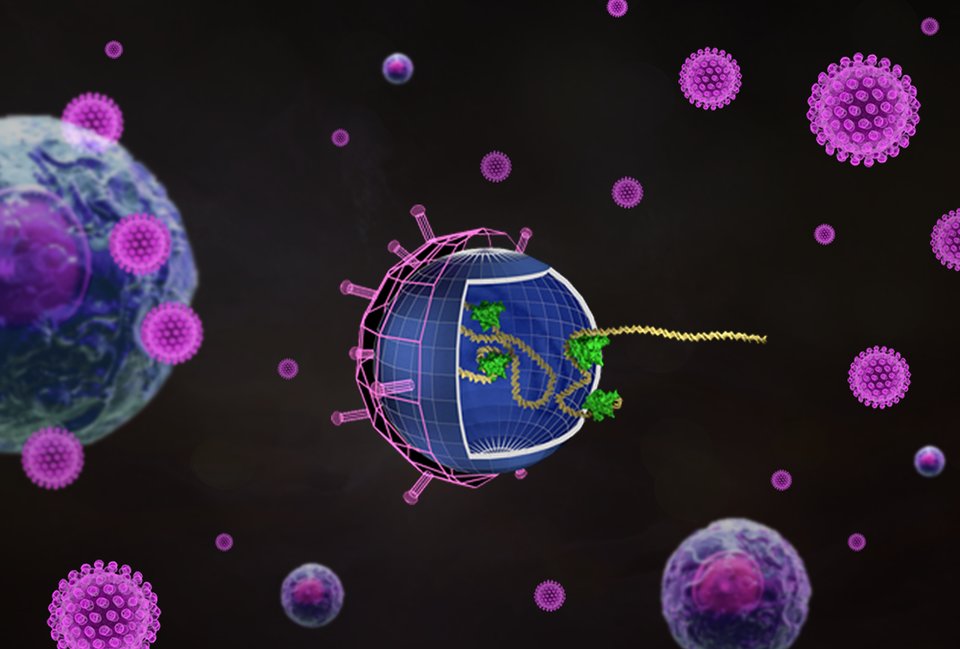
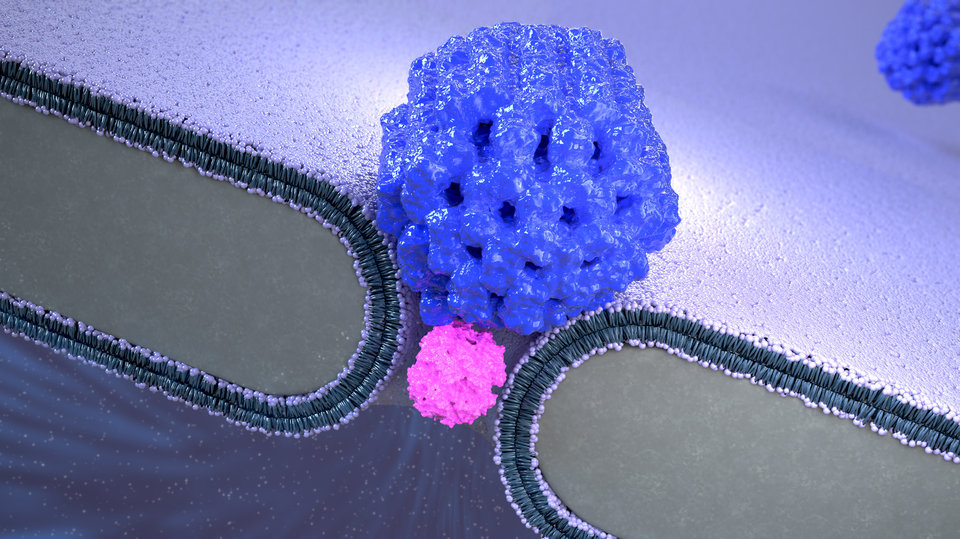
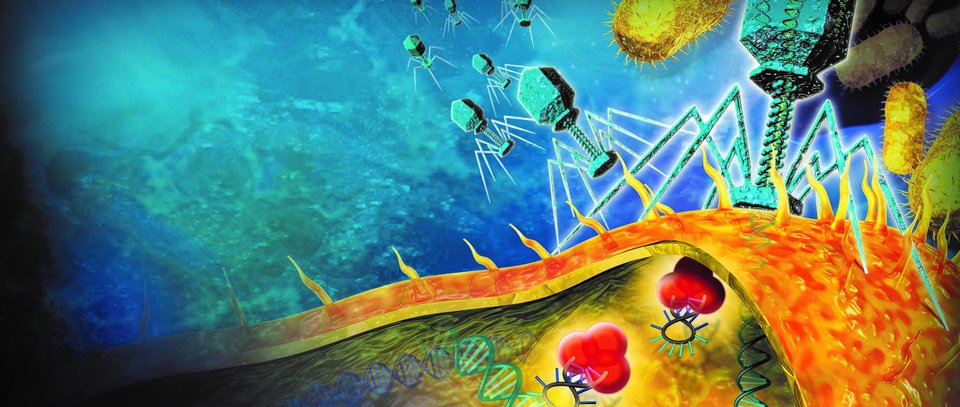
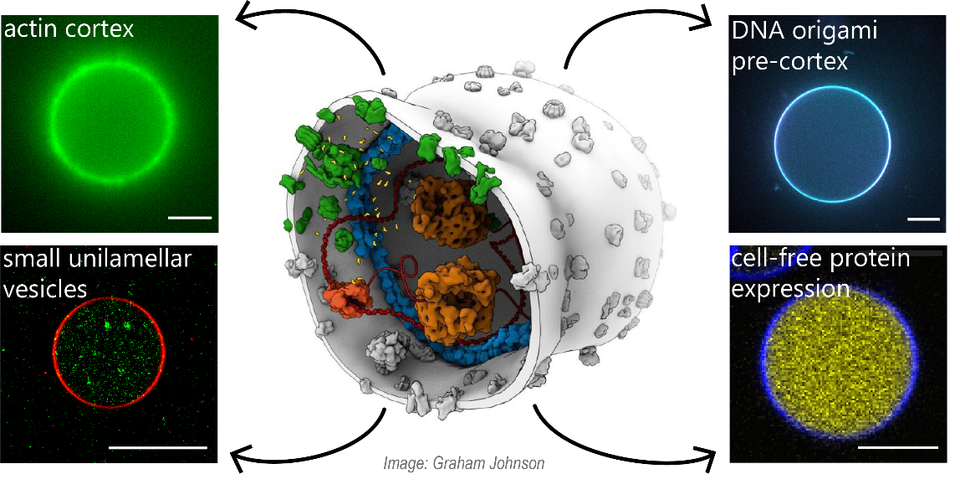
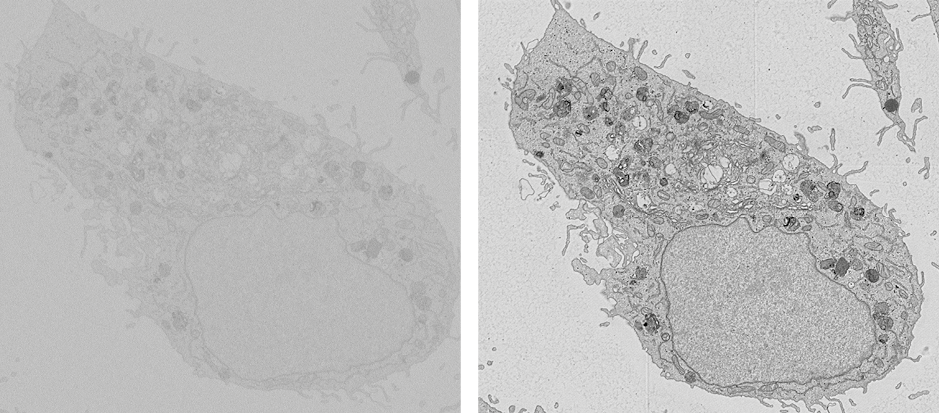
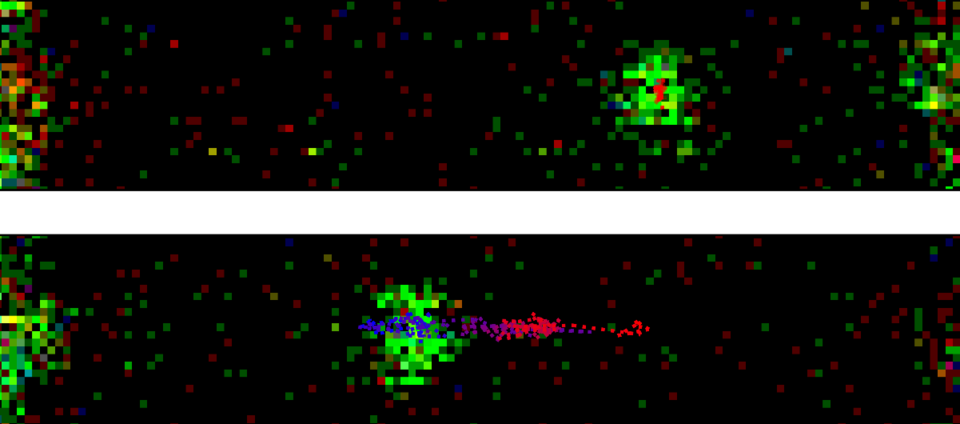
At the Faculty of Applied Sciences (AS) a lot of research is being done at the interface of technology and healthcare. Scientists at AS use yeast as a model organism for the study of degenerative brain disorders such as Alzheimer's disease, make laser set-ups and models that can visualise the blood flow in a heart chamber, study cellular processes underlying common diseases, develop new imaging techniques that can be used for hospital diagnosis, and more. This page provides an overview of the Health Technology related research carried out at AS. We distinguish four domains, in which the research of certain scientists can simultaneously take place in multiple domains.
-
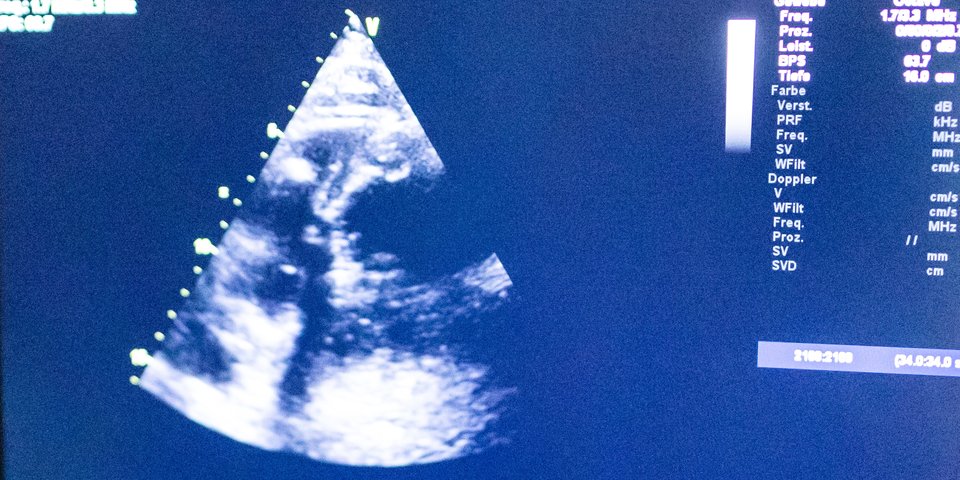
Research within this domain is aimed at making faster and better diagnoses. Think, for example, of fast and accurate imaging of diseased tissues, or the study of blood flow in order to be able to predict problems such as heart failure at an early stage.
-
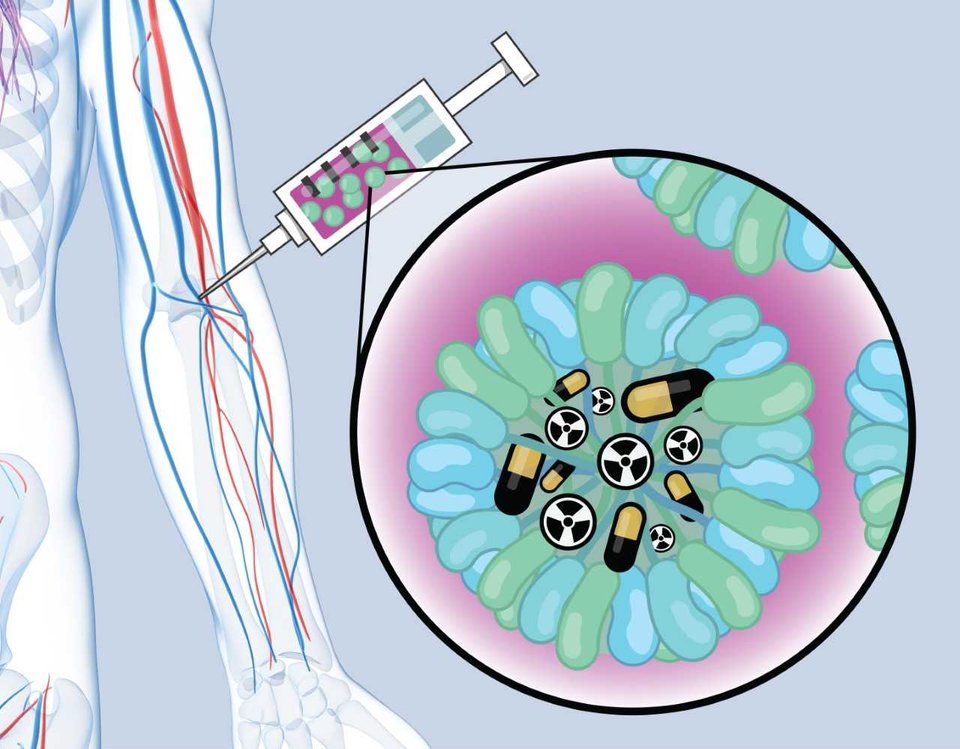
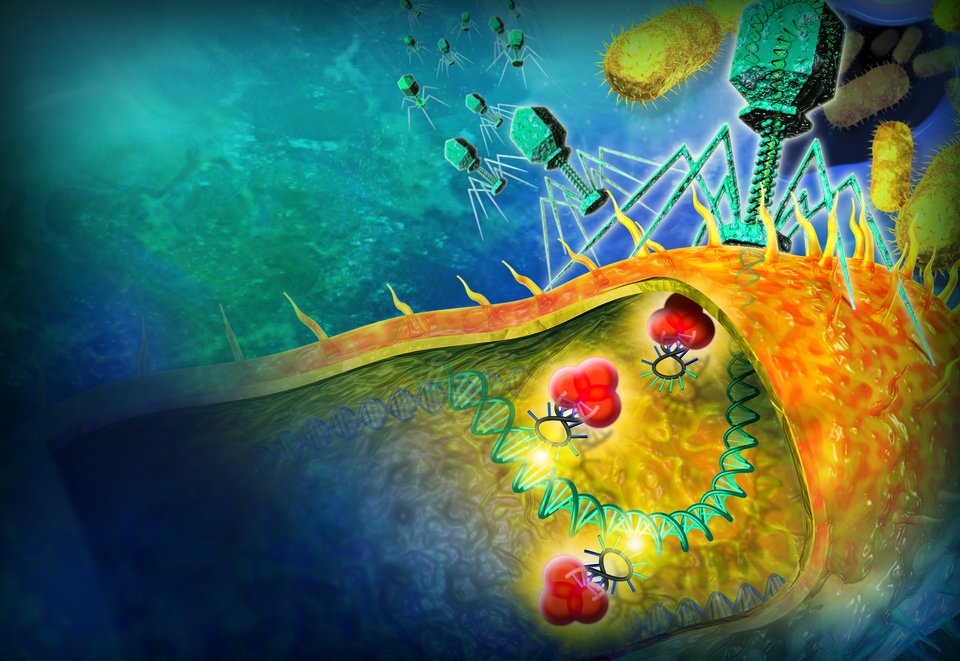
Research in this field is aimed at solving health problems. Think of personalised proton therapy that takes into account the evolution of a tumour and the changing body of a patient, or the production of antibiotics using genetically modified micro-organisms.
-
Research in this field is aimed at studying or developing processes that are essential for healthcare. Examples include fundamental research into protein folding, or the design of new industrial processes for the production of biopharmaceuticals.
-
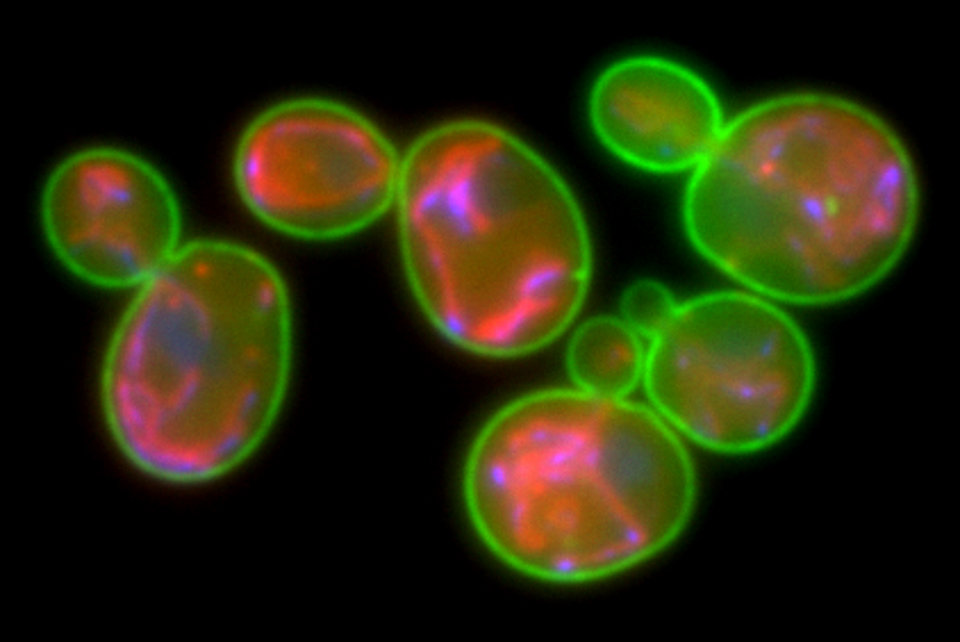
Scientific models are at the basis of many health care breakthroughs. researchers at AS are working on yeast models for research into the functioning of dementia, on animal models for answering fundamental neurological questions, and on mechanistic models that can make the industrial production of medicines more efficient.
![[Translate to English:] TU Delft Health Initiative](https://filelist.tudelft.nl/TNW/Onderzoek/T--TUDelft--healthinitiative.png)