Establishing a minimal in vitro system to study the dynamics and adaptability of a GTPase network
Summary
In vitro systems are a good way to study a broad range of protein functions in an isolated, quantitative manner. Often in in vitro studies, unexpected protein properties and function can emerge. These non-physiological functions remain hidden in vivo. Although the environment of a living cell might be quite different for the artificial in vitro environment, the properties our proteins show may have great potential in the regulation of a robust, yet adaptable cell polarity mechanism. These hidden properties could both help the cell to rescue functions of failing machinery and help the cell to adapt quickly in changing environments.
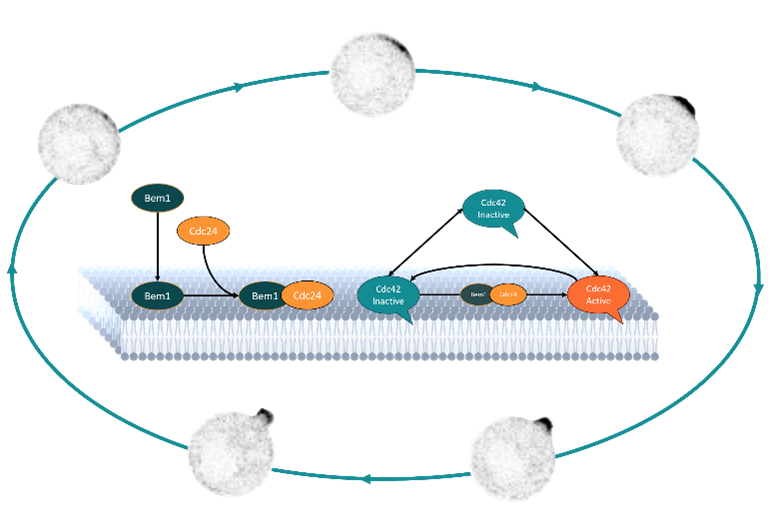
The minimal in vitro yeast polarity system we work with consists of a two-dimensional supported lipid bilayer and purified proteins. We focus on three core proteins. The polarity spot in budding yeast is defined by an accumulation of Cdc42, a GTPase. Cdc42 cell polarity is highly regulated within the cell, but also shows high adaptability. An essential protein in this regulatory network is Cdc24, this protein activates Cdc42. To bind to the membrane, Cdc24 needs scaffold protein Bem1.
The polarity proteins we study have a lot of binding partners, large disordered domains and many functional domains. This gives them a higher likelihood of finding multifunctionalities or “hidden protein functions”. We find exiting new properties like polymerization, multimerization and synergistic protein interactions.
By characterizing the hidden protein functions of key proteins in yeast cell polarity establishment, we aim to understand the mechanisms behind this fascinating, complex system.
Lab members
Dr. Sophie Tschirpke
Nynke Hettema, MSc.
Dr. Ramon van der Valk
Frank Opstal Bsc.
Ziheng Wu BSc.
Collaborators
Dr. Jos Zwanikken, Bionanoscience, TU Delft, theory and computer simulations.
Dr. Arjen Jacobi, Bionanoscience, TU Delft, electron microscopy.
Dr. Rienk Eelkema Lab, ChemE, TU Delft, organic chemistry
Dr. Martin Depken, Bionanoscience, TU Delft, kinetic theory
Papers
S Tschirpke, WKG Daalman, L Laan (2023)
The GEF Cdc24 and GAP Rga2 synergistically regulate Cdc42 GTPase cycling
bioRxiv, https://doi.org/10.1101/2023.06.26.546500
S Tschirpke, WKG Daalman, L Laan (2023)
Quantification of GTPase cycling rates of GTPases and GTPase: effector mixtures using GTPase Glo™ assays
bioRxiv, 24.568589
S Tschirpke, F van Opstal, R van der Valk, WKG Daalman, L Laan (2023)
A guide to the in vitro reconstitution of Cdc42 GTPase activity and its regulation
bioRxiv, 2023.04. 24.538075
Kim Vendel*, Sophie Tschripke*, Fayezeh Shamsi, Marileen Dogterom and Liedewij Laan (2019)
Minimal in vitro systems shed light on cell polarity.
Journal of Cell Science, 132: jcs217554 doi:10.1242 (*equal contribution)