This content is being blocked for you because it contains cookies. Would you like to view this content? By clicking here, you will automatically allow the use of cookies.

A critical look at critical materials
Read more A critical look at critical materials

From flocs to granules: new opportunities for wastewater treatment
Read more From flocs to granules: new opportunities for wastewater treatment
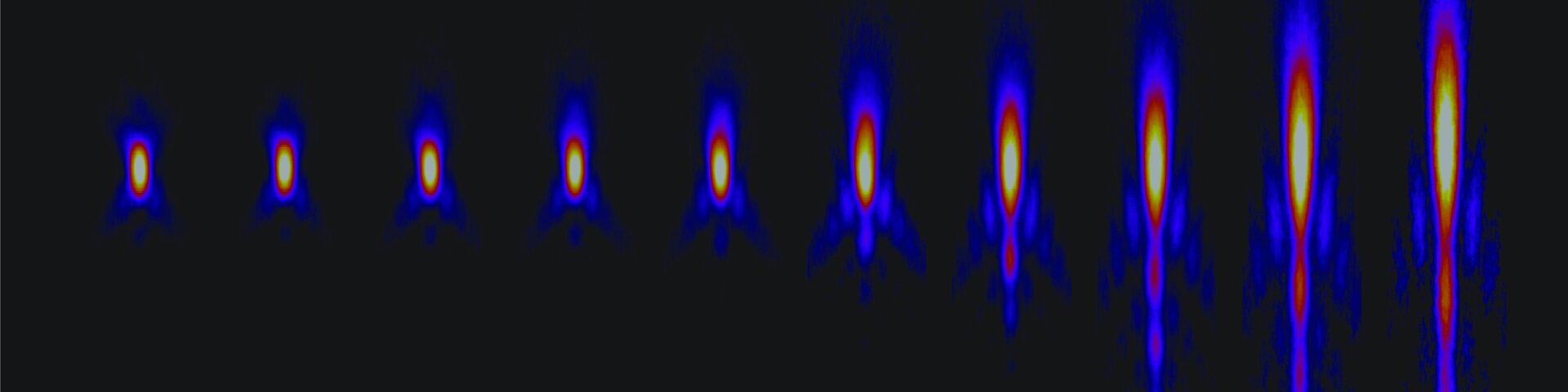
Problem in microscopy solved after decades
Read more Problem in microscopy solved after decades
Item 1
of
3