Molecular machine tears toxic protein clumps apart
How do cells disentangle proteins that are clumped together? Researchers from AMOLF in Amsterdam and Heidelberg University now show that the molecular chaperone ClpB can forcibly pull on exposed loops of protein chains, and hence extract them from protein clumps. They published their results in Nature today.
Proteins are long chains of amino acids that fold up in the body. They carry out many functions in the cell, but can only do so when they are folded properly. However, often something goes wrong causing the formation of an aggregate: a clump of proteins similar to a tangled-up ball of strings, which can cause further damage to the cell. Protein aggregation has been linked to ageing and Alzheimer’s disease, among other medical conditions.
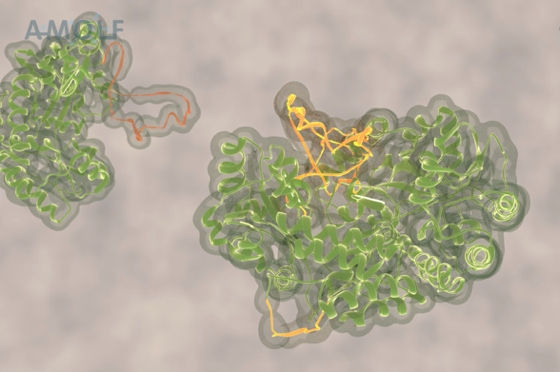
ClpB forcibly extrudes protein chain loop
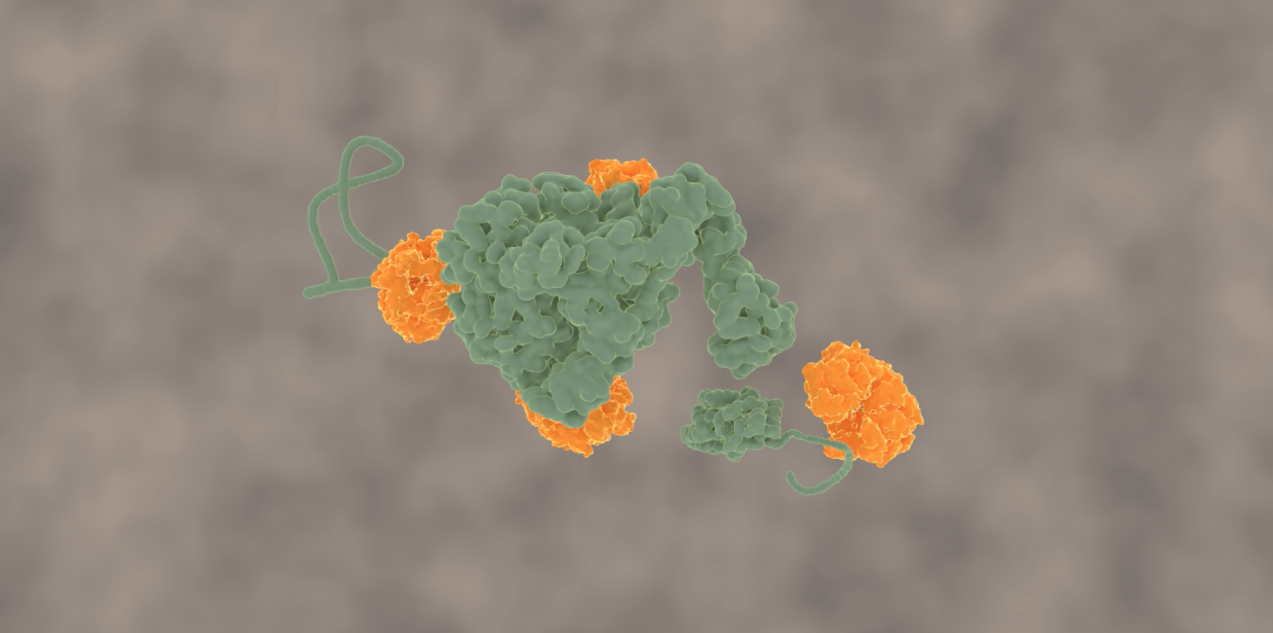
Molecular clean-up machines
Organisms have a clean-up service for these protein clumps: a group of proteins referred to as chaperones. It was known that the Hsp100 family of chaperones, of which ClpB (pronounced club B) is a member, can redissolve protein aggregates. But how they do that had never been measured directly. The central challenge is to somehow visualize all these proteins and their movements, which occur at an extremely small scale (nanometers) and are highly random.
This problem has been tackled by Mario Avellaneda and Sander Tans at the AMOLF institute in Amsterdam, in collaboration with Bernd Bukau and Axel Mogk at Heidelberg University. Their investigation has now revealed how these chaperones untangle the protein chains. Tans explains: “We found that the ring-shaped ClpB protein forcibly pulls loops of protein chains through its central pore. Such protein chain loops are present at the surface of protein clumps. However, these clumps are too large to pass through the pore. So, through this pulling action, ClpB can extract individual protein chains from the larger aggregate. Upon removal, the protein chain can fold up again and function normally. By extracting all proteins one-by-one, the chaperone can fully untangle the entire aggregate.”
Pulling with light
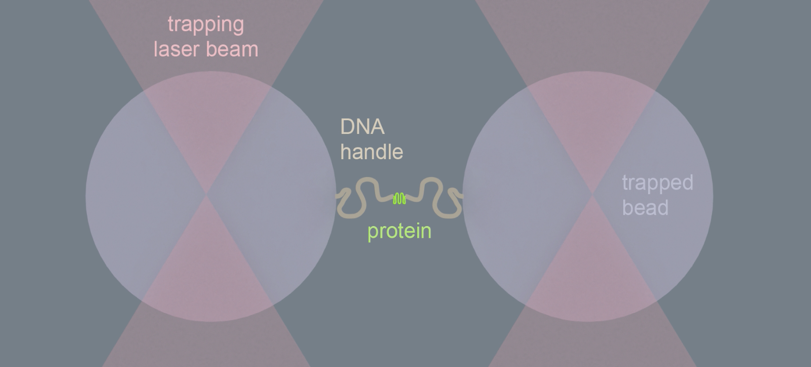
The researchers achieved this by grabbing both ends of a protein chain using optical tweezers, a technique that was awarded the Nobel prize in 2018. Optical tweezers are focused laser beams that can catch and manipulate round beads of plastic or glass. While about 1000-fold larger than proteins, one can link such beads at both protein-chain ends, and hence manipulate it very precisely. By measuring the length of the protein chain this way, the researchers could tell whether it was stretched out or compact and folded.
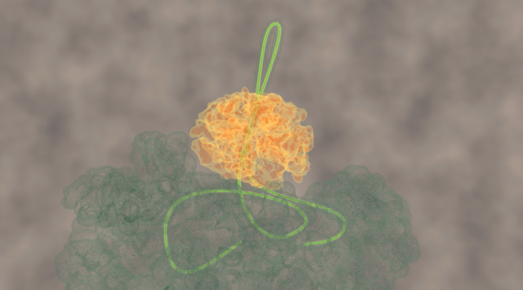
After adding the chaperone ClpB to a stretched-out chain, the researchers saw that the distance between the chain-ends was getting progressively shorter, until the ends were right next to each other. This striking observation suggested that the chain was pulled through the ring-shaped ClpB. The researchers reasoned that the protein chain ends are indeed pulled together when the protein chain is threaded through the central ClpB pore as a loop. Other experiments confirmed this idea. For instance, when the inside of the pore was modified by applying mutations, the motor-like pulling activity was abolished.
Visualizing ClpB
This extrusion of protein chains loops was never seen directly for any protein. So, to further test it, the researchers developed a different approach. The aim was to follow how ClpB moved on the protein chain, by attaching a fluorescent molecule to the chaperone. Initially they were skeptical, because these movements are smaller than the wavelength of the light that is required for the fluorescence imaging. It was thus quite a surprise when these tiny movements were indeed visible.
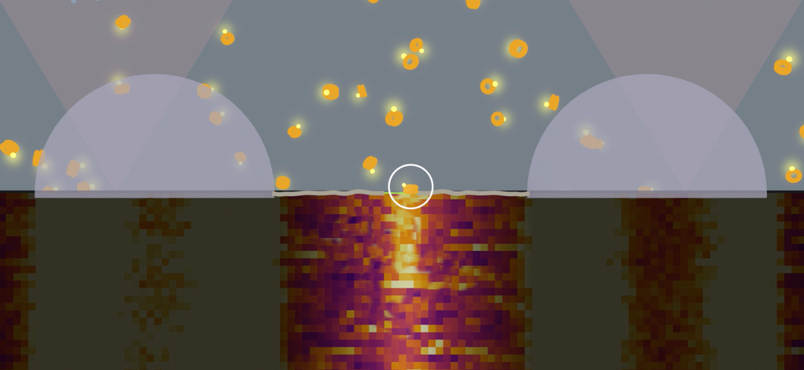
The researchers found that ClpB initially attached at a random location on the stretched protein chain. ClpB could then pull the left end of the protein chain towards itself, but also the right end, or both at the same time. Both arms of the loop could thus be pulled through the ring simultaneously. Tans: “We know how difficult it can be to stick a double thread through the eye of a needle. ClpB is able to achieve this feat using tiny actuated levers present within the pore, called pore loops. ClpB can move these levers up and down like little fingers, using the ATP fuel molecules that cells also produce. In turn this allows an amino acid chain to be pushed through the ring. Exactly how these actuated levers work is still unclear. However, we discovered that they work in bursts: first several levers push consecutively, then they pause, and then they push again, et cetera. The pause is probably needed to allow new ATP molecules to bind.”
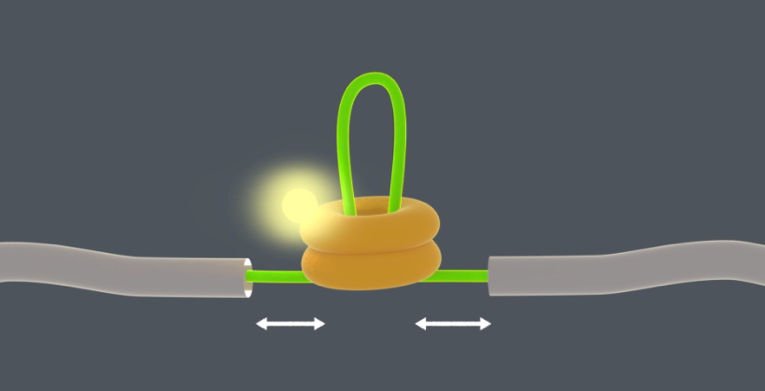
Fast and efficient
There were a number of contradicting theories on how these ClpB disaggregases works, says Tans: “One of the ideas was that many ClpB bind the sticky parts of the aggregate in random manner, a bit like soap molecules dissolving fat. Other studies suggested that at most one segment of a protein chain would fit in the ClpB channel, and that a loop would not fit for that reason. We have now demonstrated that ClpB pulls forcibly. The chaperone works like a motor that unravels aggregated proteins by pulling at exposed protein-chain loops. It keeps pulling until the protein has been completely pulled through.”
Important for people
The next step for Tans and his colleagues will be to investigate human chaperones and proteins. It is tempting to think that this research will immediately lead to a drug for Alzheimer’s disease. “In principle, you can start thinking about such things. In practice, however, it is not yet so simple,” says Tans. “We need to know better how these machines work within the cellular context before one can start working towards clinical applications. One next step is to examine how ClpB is regulated, as it can also cause damage by pulling on normal proteins.”
The research is relevant not only for unravelling protein aggregates. The fact that ClpB can pull on protein-chain loops suggests that other proteins may do the same in other cellular processes, says Tans. “For example, the cell continuously breaks down damaged proteins to make new ones – in other words, the circular economy of the cell. This can now be investigated using our methods.”
More information
Mario J. Avellaneda, Kamila B. Franke, Vanda Sunderlikova, Bernd Bukau, Axel Mogk, Sander J. Tans, Processive extrusion of polypeptide loops by a Hsp100 disaggregase, Nature 2020,
DOI: 10.1038/s41586-020-1964-y