Image analysis
BioImage Analysis - an integral part of your experiment
Acquiring good data is a prerequisite, but certainly not sufficient, for deriving meaningful information. At the KNIC facility we provide you not only with a high-end workstation and software, but also support with fitted image analysis, direct you to suitable methodologies, or point to specific optimization steps.
Your data is influenced by:
- Sample preparation (label choice)
- Imaging modality (point scanning/ light sheet/ widefield)
- Imaging parameters (bit depth/ dwell time/ pixel size/ source intensity
- Analysis parameters (threshold)
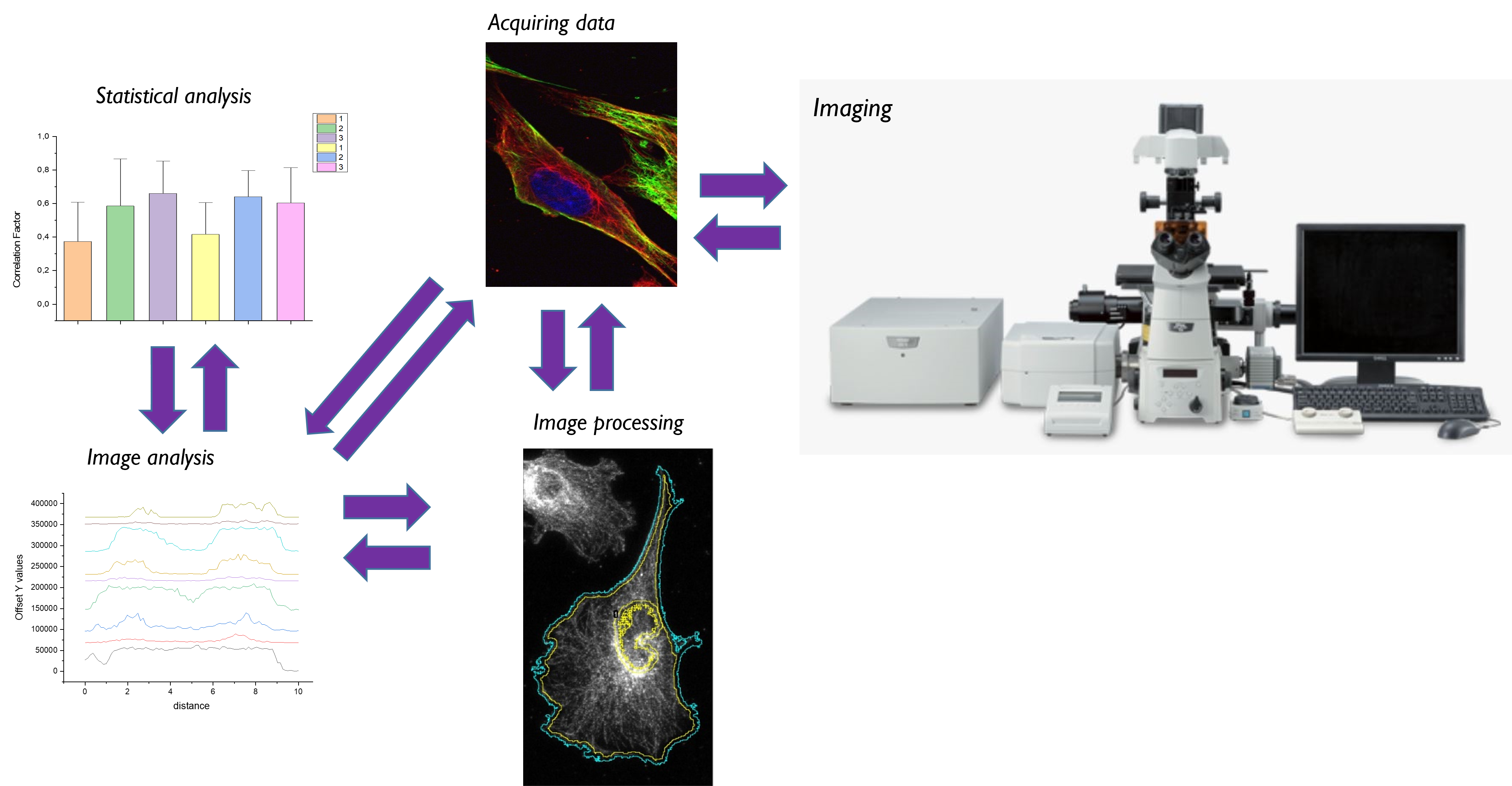
Work efficiently:
- Define your analysis - what are you aiming for?
- Morphology/Intensity/Tracking/Colocalization
- Sometimes you will need separate experiments
- Acquire a minimal dataset
- Manually verify your analysis makes sense before progressing!
Where to start?
Get familiar with sources:
Neubias, Network of European BioImage Analysts:
Training/ Algorithms/ Database/ Community
Recommended:
- Segmentation of biological samples - Intro by Jonas Ogaard
- Bioimage data analysis - Edited by Kota Miura
- Analyzing fluorescence microscopy images with ImageJ - Peter Bankhead
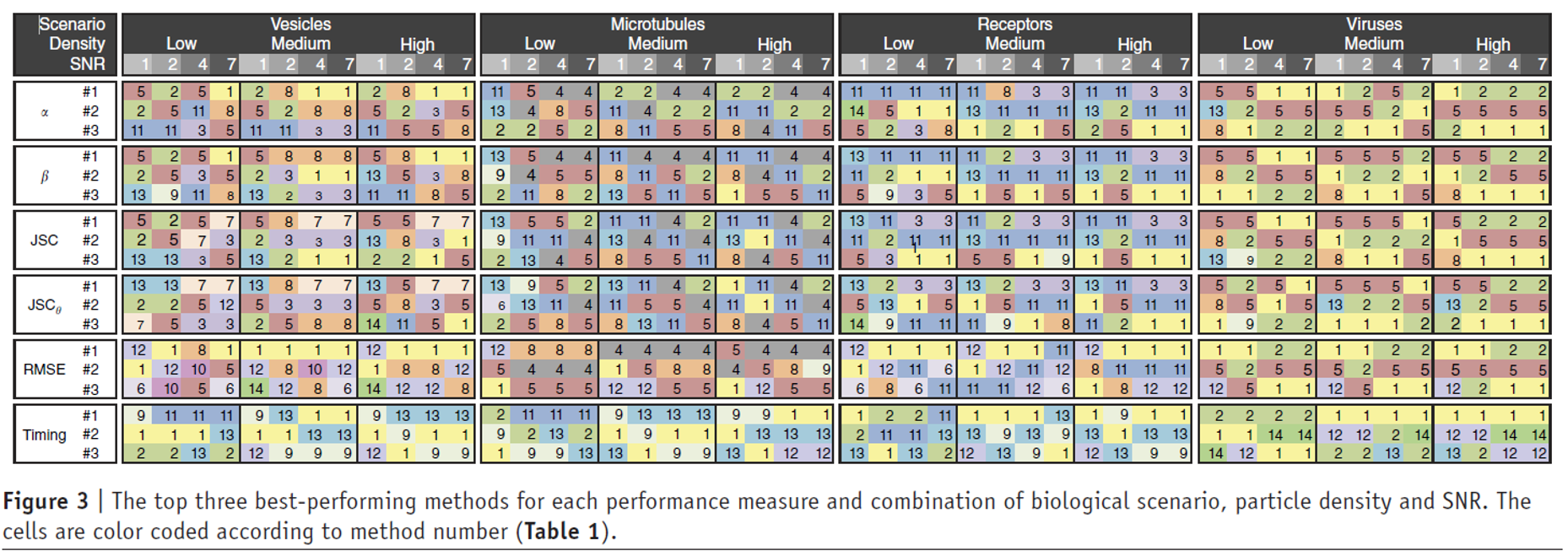
Compare methodologies
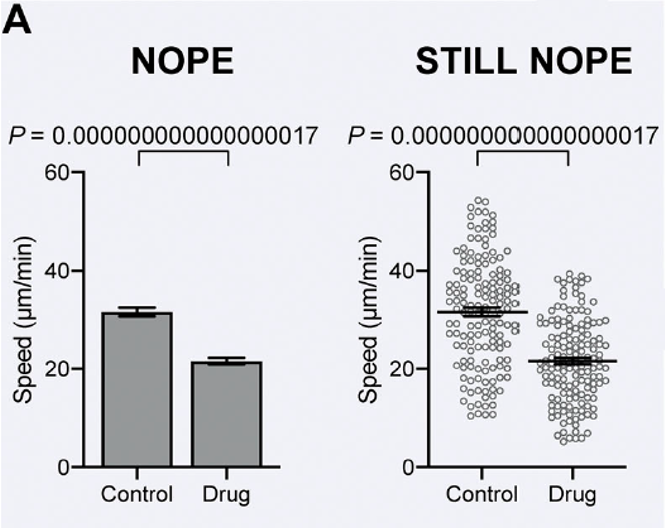
Verify significance:
SuperPlots: Communicating reproducibility and variability in cell biology
"Oh no love, you're not alone"
There are many good algorithms and open source pipelines that can fit your needs- don’t rush into developing solutions that already exist!
Check the Image.sc forum, Biii and Quarep for questions.
